| HOME | HELP | FEEDBACK | SUBSCRIPTIONS | ARCHIVE | SEARCH | TABLE OF CONTENTS |
|
| ||||||||
.gif)
.gif)

* Agricultural Research Service, USDA, Beltsville,
MD 20705-2350 Department of Bioinformatics and Computational Biology, George
Mason University, Manassas, VA 20110
Department of Bioinformatics and Computational Biology, George
Mason University, Manassas, VA 20110.gif) Division of Animal Sciences, University of Missouri, Columbia 65211
Division of Animal Sciences, University of Missouri, Columbia 65211 Center for Genetic
Improvement of Livestock, Department of Animal and Poultry Science, University
of Guelph, Ontario N1G 2W1, Canada
Center for Genetic
Improvement of Livestock, Department of Animal and Poultry Science, University
of Guelph, Ontario N1G 2W1, Canada
1 Corresponding author: George.Wiggans{at}ars.usda.gov
Nearly 57,000 single-nucleotide polymorphisms (SNP)
genotyped with the Illumina BovineSNP50 BeadChip (Illumina Inc., San
Diego, CA) were investigated to determine usefulness of the
associated SNP for genomic prediction. Genotypes were obtained for
12,591 bulls and cows, and SNP were selected based on 5,503 bulls
with genotypes from a larger set of SNP. The following SNP were
deleted: 6,572 that were monomorphic, 3,213 with scoring problems
(primarily because of poor definition of clusters and excess number
of clusters), and 3,649 with a minor allele frequency of <2%.
Number of SNP for each minor allele frequency class (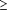 2%) was fairly uniform (777 to
1,004). For 5 contiguous SNP assigned to chromosome 7, no bulls were
heterozygous, which indicated that those SNP are actually on the
nonpseudoautosomal portion of the X chromosome. Another 178 SNP that
were not assigned to a chromosome but that had many fewer
heterozygotes than expected were also assigned to the X chromosome.
Existence of Hardy-Weinberg equilibrium was investigated by comparing
observed with expected heterozygosity. For 11 SNP, the observed
percentage of heterozygous individuals differed from the expected by
>15%; therefore, those SNP were deleted. For 2,628 SNP, the
genotype at another SNP was highly correlated (i.e., genotypes were
identical for >99.5% of bulls), and those were deleted. After
edits, 40,874 SNP remained. A parent–progeny conflict was declared
when the genotypes were alternate homozygotes. Mean number of
conflicts was 2.3 when pedigree was correct and 2,411 when it was
incorrect. The sire was genotyped for >93% of animals. Maternal
grandsire genotype was similarly checked; however, because alternate
homozygotes could be valid, a conflict threshold of 16% was used to
indicate a need for further investigation. Genotyping consistency
was investigated for 21 bulls genotyped twice with differences
primarily from SNP that were not scored in one of the genotypes.
Concordance for readable SNP was extremely high (99.96–100%).
Thousands of SNP that were polymorphic in Holsteins were monomorphic
in Jerseys or Brown Swiss, which indicated that breed-specific
SNP sets are required or that all breeds need to be considered
in the SNP selection process. Genotypes from the Illumina BovineSNP50
BeadChip are of high accuracy and provide the basis for genomic
evaluations in the United States and Canada.
2%) was fairly uniform (777 to
1,004). For 5 contiguous SNP assigned to chromosome 7, no bulls were
heterozygous, which indicated that those SNP are actually on the
nonpseudoautosomal portion of the X chromosome. Another 178 SNP that
were not assigned to a chromosome but that had many fewer
heterozygotes than expected were also assigned to the X chromosome.
Existence of Hardy-Weinberg equilibrium was investigated by comparing
observed with expected heterozygosity. For 11 SNP, the observed
percentage of heterozygous individuals differed from the expected by
>15%; therefore, those SNP were deleted. For 2,628 SNP, the
genotype at another SNP was highly correlated (i.e., genotypes were
identical for >99.5% of bulls), and those were deleted. After
edits, 40,874 SNP remained. A parent–progeny conflict was declared
when the genotypes were alternate homozygotes. Mean number of
conflicts was 2.3 when pedigree was correct and 2,411 when it was
incorrect. The sire was genotyped for >93% of animals. Maternal
grandsire genotype was similarly checked; however, because alternate
homozygotes could be valid, a conflict threshold of 16% was used to
indicate a need for further investigation. Genotyping consistency
was investigated for 21 bulls genotyped twice with differences
primarily from SNP that were not scored in one of the genotypes.
Concordance for readable SNP was extremely high (99.96–100%).
Thousands of SNP that were polymorphic in Holsteins were monomorphic
in Jerseys or Brown Swiss, which indicated that breed-specific
SNP sets are required or that all breeds need to be considered
in the SNP selection process. Genotypes from the Illumina BovineSNP50
BeadChip are of high accuracy and provide the basis for genomic
evaluations in the United States and Canada.
Key Words: genomic prediction • genotyping • single-nucleotide polymorphism
| HOME | HELP | FEEDBACK | SUBSCRIPTIONS | ARCHIVE | SEARCH | TABLE OF CONTENTS |