GENETIC EVALUATION SYSTEMS IN THE UNITED STATES
Wiggans, G.R.
Animal Improvement Programs Laboratory, Agricultural Research Service, U.S. Department of Agriculture, Beltsville, MD 20705-2350, USA
(This text was excerpted from a presentation at the 32nd International Symposium, Animal Production: Advances in Technology, Accuracy, and Management, September 29 - October 1, 1997, Milan, Italy)
Abstract: The genetic evaluation system for dairy animals in the United States applies a state-of-the-art animal model to produce evaluations that are used to achieve a high rate of genetic progress. The system is modified frequently to meet the needs of the U.S. dairy industry and to exploit advances in computer technology and evaluation methodology. The evaluations and supplemental information are distributed widely, primarily through industry cooperators and the internet. The current animal model, which was implemented in 1989, enables all relatives of each cow with lactation records to contribute to her evaluation and to the evaluation of her sire. Genetic groups are defined for unknown parents to allow recognition of the improvement in genetic merit over time and of the differences between unknown sires and dams. Records from the first five lactations are included using a repeatability model. A first lactation is required for a record to affect evaluations of relatives. Changes since 1989 include: (1) the use of records from later herds for cows that change herds, (2) an accounting for the reduced genetic variance of projected records, (3) an adjustment for heterogeneous variance, (4) the inclusion of age and parity in the model as well as the multiplicative adjustment of records prior to analysis, (5) an accounting for inbreeding when forming the inverse of the relationship matrix, (6) the incorporation of information from Canadian genetic evaluations, (7) the calculation of genetic evaluations for productive life (longevity) and somatic cell score (mastitis resistance), and (8) the inclusion of unsupervised records after additional editing to remove less reliable data.
U.S. Dairy Cattle Population
On January 1, 1997, about 4.5 of the 9.3 million cows in the United States were enrolled in test plans of the National Cooperative DHI Program (Wiggans, 1997a). Approximately 2.5 million of these cows met requirements to contribute data to genetic evaluations. Of the cows tested, 93% were Holsteins, and 3% were Jerseys. The average herd size for the tested population was 109 cows. The tested population averaged 8578 kg of milk, 321 kg of fat, and 278 kg of protein for 1996 (Wiggans, 1997b). Nearly 15% of the cows (about 5 % of the herds) were milked three times a day, and milk from 85% of the cows was analyzed for somatic cell count.
Milk Recording
Data collection on farms and determination of milk components is managed by 32 DHI affiliates. Geographical coverage by individual affiliates ranges from a county to several states. These affiliates contract with dairy records processing centers to manage data. Seven processing centers prepare reports for producers and forward data to the Animal Improvement Programs Laboratory, part of the U.S. Department of Agriculture’s Agricultural Research Service, for use in the calculation of genetic evaluations.
As cost pressures have forced dairy producers to become more efficient and as herd size has increased, the milk-recording program has evolved. In an effort to maintain the numbers of cows and farms that participate in milk recording, considerable flexibility has been allowed in data collection, which varies by the amount of supervision provided by the DHI affiliate, the amount of component sampling, and the frequency of milk recording. Only 30% of cows have all milkings on test day weighed and sampled. The most popular data collection method (60%) is weighing and sampling only one milking on test day. The remaining 10% of cows have all milkings weighed on test day, but only one milking is sampled so that the labor and disruption from the sampling process are reduced.
Electronic milk recording is used for some herds to record daily milk weights. An average of these weights is reported as the test-day milk yield. Currently, component samples are required for data to be included in genetic evaluations; therefore, electronic meters have not eliminated all the labor required for milk recording.
Dairy producers can arrange for a wide range of data collection schedules. For example, they can alternate supervised and unsupervised tests. Because of this greater variety, a herd profile and a data collection rating were developed to enable users of the data to know how the data were collected. Previously, standard test plans were defined for this purpose (Benson, 1985). The herd profile indicates when the herd was tested and if the tests were supervised (example shown at http://www.dhia.org/lib04.htm). The herd profile also reports outliers among the herd’s data. The data collection rating is based on the expected correlation between lactation records with the information recording characteristics of a particular herd and lactation records calculated from 10 equally spaced tests and samples. These new resources greatly reduce the rule-enforcement role of milk recording agencies and place the responsibility on users to decide if data are suitable.
Until February 1997, only data from supervised test plans were included in evaluations. Unsupervised data now are included if the herd meets additional restrictions. Supplementary restrictions imposed on those data include limits on the proportion of the herd with missing identification, the difference between milk shipped and the sum of the test-day weights, and the number of outliers (details at http://aipl.arsusda.gov/reference/useos.htm).
Yield Evaluations
Genetic evaluations for yield traits are calculated at the Animal Improvement Programs Laboratory from data received from dairy records processing centers (Wiggans and VanRaden, 1993). Lactation records from calvings in 1960 and later are included in evaluations. Incoming data are subject to many checks for reasonable values and consistency with existing data (Norman et al., 1994) and are adjusted for calving season, length of lactation, and number of milkings per day prior to analysis. Evaluations are calculated four times a year with an animal model (VanRaden et al., 1995; Wiggans et al., 1988; Wiggans and VanRaden, 1989), which initially was implemented in 1989. The current animal model describes a cow’s lactation as the sum of the effects of her management group (m), genetic merit (a), permanent environment (p), interaction of her herd and sire (c), calving age and parity (v), and unexplained residual (e):
y = m + a + p + c + v + e.
The genetic merit (or animal effect) is an animal’s breeding value; the predicted transmitting ability (PTA) reported to the dairy industry is half the animal’s breeding value. The inclusion of an effect for herd-sire interaction limits the impact on a bull's evaluation from daughters in a single herd.
For milk, fat, and protein evaluations, the proportions of differences between records attributable to the various effects (variance components) are assumed to be 30% for genetics (heritability), 15% for permanent environment, 10% for herd-sire interaction (environmental correlation), and 45% for unexplained residual (Van Tassell et al., 1997). The correlation between repeated records is 0.55 and is the sum of heritability, the environmental correlation, and the variation of permanent environment. Prior to August 1997, heritability for U.S. genetic evaluations for yield traits had been assumed to be 25%. The increase in heritability placed more emphasis on an animal’s performance relative to information from its relatives. Therefore, to limit the impact on evaluations from large deviations from management group averages, management group deviations were limited to ±4 herd-year standard deviations. The increase in heritability has been shown to improve the ability of evaluations based on first-crop daughter records to predict later daughter performance (Van Tassell et al., 1997).
The current genetic base was established by setting the average evaluation of cows born in 1990 to 0. The genetic base in the United States is updated every 5 years. The next base change will be in the year 2000, when cows born in 1995 will become the base population.
Lactation records after fifth parity are not included in any genetic evaluations. A lactation record for first parity is required for a cow’s data to contribute to the evaluations of her relatives. For cows with no reported first lactation but born in the preceding 10 years, an evaluation is calculated that includes the later lactation records, but this evaluation does not contribute information to evaluations of her relatives (Wiggans and VanRaden, 1990). All records of a cow across herds contribute to the estimation of permanent environment and herd-sire interaction effects for her first herd.
Genetic variation is stabilized in two ways. Deviations from incomplete records are expanded based on the number of tests (VanRaden et al., 1991), and all records are adjusted based on phenotypic variation estimated on a herd-year basis (Wiggans and VanRaden, 1991). Because of the variability of these estimates of phenotypic variation, they are smoothed with values from adjacent years and a regional value.
An effect for age-parity (Wiggans and VanRaden, 1994) is included in the model to account for residual effects not removed by multiplicative adjustments of records prior to analysis. Estimates of genetic trend have been found to be quite sensitive to age adjustments.
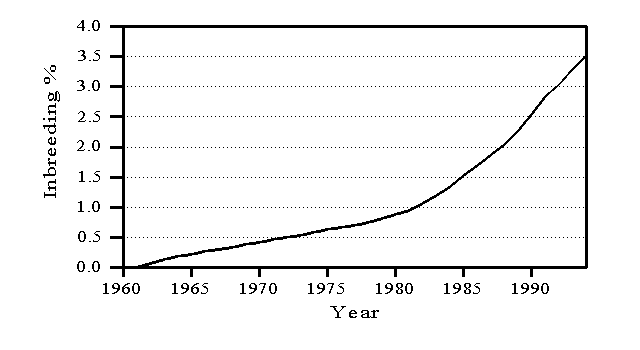
Inbreeding is defined relative to animals born before 1960, which are considered unrelated, and measures the likelihood that an animal will inherit the same allele from an ancestor common to both parents. Inbreeding reduces genetic variation and, therefore, is considered when forming the inverse of the relationship matrix (VanRaden, 1992; Wiggans et al., 1995). Average inbreeding has increased each year as shown in Figure 1. When an animal has an unknown parent, the animal’s percentage of inbreeding is assumed to be the average for its birth year. Inbreeding can be reduced by using mating plans that consider relationships among animals. No adjustment currently is made to genetic evaluations for inbreeding depression because such an adjustment would increase the evaluations of bulls that are likely to be related to their mates and, therefore, would contribute to an increase in inbreeding (Lawlor et al., 1993).

Figure 1. Trend in inbreeding for U.S. Holsteins.
With the globalization of dairy cattle breeding, some bulls are evaluated that have Canadian sires or dams. To represent the pedigree contribution of such bulls appropriately, dam evaluations are requested from Canada, and sire evaluations from the previous International Bull Evaluation Service (Interbull) evaluation are collected. Dam evaluations are 3 months old, and sire evaluations are 3 to 6 months old. These dam and sire evaluations are used to update the parent contribution to a bull’s evaluation when the information from outside the United States has significantly higher reliability (accuracy). This procedure, which is based on the system of combining evaluations reported by Wiggans et al. (1992), is applied after iteration and only for bull evaluations. Its primary benefit is greater accuracy for parent averages of young bulls. To extend this practice to bull dams from other countries would require a convenient source of dam evaluations; Interbull is a possible central access point for acquiring dam evaluations in the future.
In addition to PTA, several other statistics are reported to the U.S. dairy industry to aid in understanding how an evaluation is generated (VanRaden and Wiggans, 1991):
Genetic Trend for Yield
Average breeding value of cows by birth year is a common measure of genetic trend and an indication of the success of a national breeding program. However, trend estimates can be affected by the evaluation model and the adjustments for age effects (Wiggans and VanRaden, 1994). Figure 2 shows the trend in breeding values for protein yield of U.S. Holstein cows. The earliest birth date included is 1975 because relatively few cows had protein information collected before then. For cows born in 1993, the latest year with complete data, the average increase in breeding value was 4.2 kg of protein, which is 1.3% of average yield.
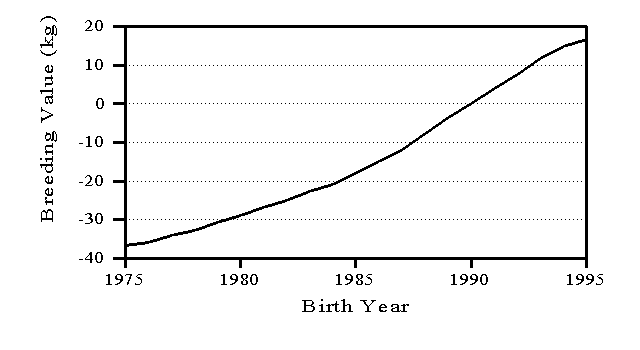
Figure 2. Trend in breeding values for protein yield for U.S. Holsteins.
Productive Life Evaluations
Productive life (PL) (VanRaden and Klaaskate, 1993) in the United States is defined as the number of months in milk (with a maximum of 10 months per lactation) until the cow is 84 months old. For cows that have not completed their productive lives, the months in milk are projected. Cows less than 30 months old are not evaluated. Information from cows born in 1960 or later is included. Because there is only one observation per cow, no permanent environmental variation is assigned, and no age-parity effect is included. The variance components as a percentage of phenotypic variation are 8.5% for heritability, 5% for herd-sire interaction, and 86.5% for unexplained residual. The low heritability for PL results in greater emphasis on parent average in the evaluation and lower reliabilities for bulls. The same computer programs are used for the analysis; therefore, most other aspects of PL evaluations (VanRaden and Wiggans, 1995) are the same as for yield evaluations.
To add accuracy for bulls, information on PL from type traits is added (Weigel, 1996). The Holstein Association USA (Brattleboro, Vermont) combines evaluations for PL, milk and fat yields, and linear type traits to calculate an approximate multitrait evaluation. This enhancement currently is applied only for Holstein bulls and has its greatest benefit for bulls with a type evaluation but that have a yield evaluation that is based on daughters that are too young to contribute PL information.
Somatic Cell Score Evaluations
Somatic cell score (SCS) evaluations were implemented in 1994 in the United States (Schutz, 1994). An SCS is a log2 transformation of somatic cell count and is related to mastitis incidence. The transformation makes the values nearly normally distributed and has the simple interpretation that each increase of 1 is a doubling of the somatic cell count. As with yield, lactation averages for up to the first five lactations are included.
Because expected SCS changes over the lactation, short records are adjusted to a complete lactation equivalent. Records are multiplicatively adjusted for age (Schutz et al., 1995). The model for yield traits is applied. The variance components as a percentage of phenotypic variation are 10% for heritability, 20% for permanent environment, 5% for herd-sire interaction, and 65% for unexplained residual.
In contrast to other yield-related traits, the selection goal for SCS is downward. Because of the large costs associated with mastitis but relatively low heritability of SCS, there was concern that the usual scale for reporting evaluations, which is centered on 0, might lead to a refusal to use animals with an evaluation higher than 0. To avoid this problem, SCS evaluations are centered around breed average, which is 3.2 for Holsteins.
Type Evaluations
Genetic evaluations of linear type traits for Holsteins are calculated by Holstein Association USA. A multitrait animal model (Misztal et al., 1993) is applied with adjustment for heterogeneous variances (Weigel and Lawlor, 1994). A canonical transformation is used to create uncorrelated traits for analysis. Because the data include more than one scoring per cow, a permanent environmental effect is included. Misztal et al. (1995) developed a method to perform canonical transformation with the additional random effect of permanent environment.
For other breeds, the Animal Improvement Programs Laboratory has calculated linear type evaluations with a single-trait sire model (Norman et al., 1979). An animal model system is being developed that does not require all traits to be measured for all animals (Gengler et al., 1997). This relaxation allows new traits to be included in the multitrait system and to gain accuracy from the information provided by correlated traits. The animal model system is expected to be implemented in February 1998.
Calving Ease Evaluations
The National Association of Animal Breeders funds the analysis of calving difficulty. Using a categorical model (Berger, 1994), calving ease evaluations are calculated for bulls at Iowa State University. Evaluations are reported as the expected percentage of difficult births for first-calf heifers giving birth to a bull calf during the winter. These evaluations have been particularly helpful in promoting the use of AI dairy bulls with virgin heifers.
Economic Indexes
Combining the many evaluations available for yield and type traits into an economic index is helpful in promoting appropriate use of the genetic information. Without such indexes, dairy producers are likely to set minimum values for specific traits and, thus, overlook some bulls that are outstanding for traits of primary economic importance. The Animal Improvement Programs Laboratory calculates a net merit dollars (NM$) index that combines evaluations for milk, fat, protein, PL, and SCS (VanRaden and Wiggans, 1995). This index includes a milk-fat-protein dollars (MFP$) index that combines yield evaluations based on projected prices. The MFP$ formula to be used until the year 2000 is:
MFP$ = $0.031 (PTA milk pounds) + $0.80 (PTA fat pounds) + $2.00 (PTA protein pounds).
The NM$ is based on a relative weighting of 10:4:-1 for yield:PL:SCS:
NM$ = 0.7 (MFP$) + $11.30 (PTA PL) - $28.22 (PTA SCS - breed average SCS).
The NM$ was developed with the implementation of PL and SCS evaluations to assist dairy producers and breeders to use information for the new traits appropriately.
Holstein Association USA calculates a type-production index (TPI) (Holstein Association, 1997):
TPI = [ 3 (PTA protein pounds/19) + 1 (PTA fat pounds/22.5) + 1 (PTA type/.7) + .65 (udder composite/.8) + .35 (feet and legs composite/.85) ] 50 + 576.
The TPI goal is to include type traits that impact profitability of the animal. This index heavily emphasizes PTA protein with no direct weight on PTA milk. However, selection for fat and protein yields tends to increase milk yield as well.
Distribution of Genetic Information
Genetic evaluations are released in February, May, August, and November. Information from a minimum of 10 daughters is required for release of a bull evaluation. Numbers of AI bulls with U.S. evaluations distributed are shown in Figure 3 by birth year. To minimize the size of files with cow evaluations, only cows born in the preceding 15 years and dams born in the preceding 20 years have their U.S. evaluations distributed (Figure 4). In both Figures 3 and 4, the counts drop for recent years because most animals born in those years have not been evaluated yet.
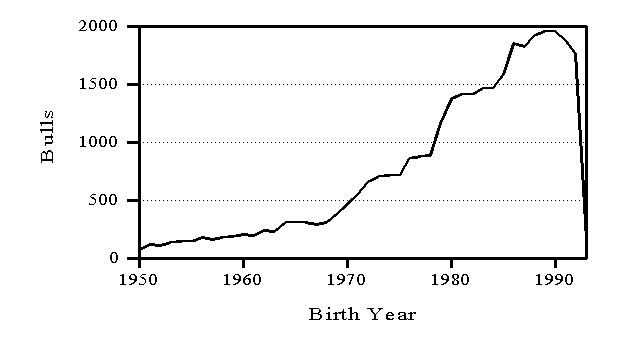
Figure 3. Numbers of AI bulls with evaluations distributed in the United States by birth year of bull.
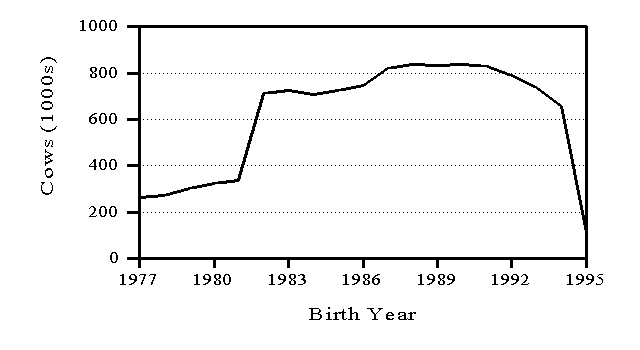
Figure 4. Numbers of cows with evaluations distributed in the United States by birth year of cow.
To minimize costs associated with upkeep and shipment of magnetic media, the internet is the primary method of distribution for U.S. genetic evaluations. Evaluation data are provided in several files and formats to accommodate the needs of users. Most evaluation files (both general access and organization-specific) are available for downloading in a compressed, password-protected format before release day to reduce electronic transfer congestion on release day. The password to access the files is provided on release day. Password-protected sites are provided to individual AI organizations for files of bull and daughter evaluations. Other bull owners are mailed reports for their bulls. Large files of cow evaluations are copied to magnetic tape and delivered by overnight courier services to some breed registry societies and dairy records processing centers.
Updated evaluations from Interbull are made available in February and August on release day after determining whether the U.S. or Interbull evaluation is to be considered official in the United States. An Interbull evaluation is official if the U.S. evaluation has a reliability of less than 80% and the Interbull reliability is greater than the U.S. reliability by 5% or more (VanRaden et al., 1997).
All genetic evaluation information calculated by the Animal Improvement Program Laboratory is accessible through a web site (http://aipl.arsusda.gov). Individual animal queries are possible, and files with subsets of bull and cow evaluations can be downloaded. Access to additional query functions enables breed registry societies and dairy records processing centers to list all the cows in a cow’s management group, to display a four-generation pedigree for an animal and its ancestors, and to select evaluation records for groups of cows or for all the cows in groups of herds. These on-line tools enable cooperators to investigate evaluations of individual cows and to acquire genetic information on the cows of new clients. Password-protected areas allow exchange of files specific to cooperating agencies including breed societies, processing centers, and AI organizations. Individual dairy producers also can obtain genetic information from dairy records processing centers and breed registry societies.
Acknowledgments
The author thanks T.J. Lawlor, Holstein Association USA; N. Gengler, Chargé de Recherches, Fonds National Belge de la Recherche Scientifique; and J.C. Philpot and S.M. Hubbard, Animal Improvement Programs Laboratory, Agricultural Research Service, U.S. Department of Agriculture, for manuscript review.
References
Benson R.H. The NCDHIP record plans. National Cooperative Dairy Herd Improvement Program Handbook, Fact Sheet A-4. Extension Service, U.S. Department of Agriculture. Washington, DC. 1985.
Berger P.J. Genetic prediction for calving ease in the United States: data, models, and uses by the dairy industry. J. Dairy Sci. 1994; 77: 1146-1153.
Gengler N., Wiggans G.R., Wright J.R., Norman H.D., Wolfe C.W. Application of canonical transformation with missing values to multitrait evaluation of Jersey type. J. Dairy Sci. 1997; 80: 2563-2571.
Holstein Association USA. Holstein Type-Production Sire Summaries, February 1997. Holstein Association. Brattleboro, Vermont.
Lawlor T.J. Jr., Weigel K.A., Misztal I. Implications of incorporating inbreeding information into animal model evaluations for type (abstract). J. Dairy Sci. 1993; 76(Suppl. 1): 292 (P435).
Misztal I., Lawlor T.J., Short T.H. Implementation of single- and multiple-trait animal models for genetic evaluation of Holstein type traits. J. Dairy Sci. 1993; 76: 1421-1432.
Misztal I, Weigel K., Lawlor T.J. Approximation of estimates of (co)variance components with multiple-trait restricted maximum likelihood by multiple diagonalization for more than one random effect. J. Dairy Sci. 1995; 78: 1862-1872.
Norman H.D., Cassell B.G., Dickinson F.N., Wright E.E. Sire evaluation for conformation of Jersey cows. J. Dairy Sci. 1979; 62:1914-1921.
Norman H.D., Waite L.G., Wiggans G.R., Walton L.M. Improving accuracy of the United States genetics database with a new editing system for dairy records. J. Dairy Sci. 1994; 77: 3198-3208.
Schutz M.M. Genetic evaluation of somatic cell scores for United States dairy cattle. J. Dairy Sci. 1994; 77: 2113-2129.
Schutz M.M., VanRaden P.M., Wiggans G.R., Norman H.D. Standardization of lactation means of somatic cell scores for calculation of genetic evaluations. J. Dairy Sci. 1995; 78: 1843-1854.
VanRaden P.M. Accounting for inbreeding and crossbreeding in genetic evaluation of large populations. J. Dairy Sci. 1992; 75: 3136-3144.
VanRaden P.M., Klaaskate E.J.H. Genetic evaluation of length of productive life including predicted longevity of live cows. J. Dairy Sci. 1993; 76: 2758-2764.
VanRaden P.M., Wiggans G.R. Derivation, calculation, and use of national animal model information. J. Dairy Sci. 1991; 74: 2737-2746.
VanRaden P.M., Wiggans G.R. Productive life evaluations, accuracy, and economic value. J. Dairy Sci. 1995; 78: 631-638.
VanRaden P.M., Wiggans G.R., Ernst C.A. Expansion of projected lactation yield to stabilize genetic variance. J. Dairy Sci. 1991; 74: 4344-4349.
VanRaden P.M., Wiggans G.R., Norman H.D., Powell, R.L. Changes in USDA-DHIA genetic evaluations (February 1997). AIPL Res. Rpt. 1997; CH7. [http://aipl.arsusda.gov/reference/changes/chng972.pdf].
VanRaden P.M., Wiggans G.R., Powell R.L., Norman H.D. Changes in USDA-DHIA genetic evaluations (January 1995). AIPL Res. Rpt. 1995; CH3. [http://aipl.arsusda.gov/reference/changes/chng951.pdf].
Van Tassell C.P., Wiggans G.R., VanRaden, P.M., Norman, H.D. Changes in USDA-DHIA genetic evaluations (August 1997). AIPL Res. Rpt. 1997; CH9. [http://aipl.arsusda.gov/reference/changes/chng978.pdf].
Weigel K.A. Use of correlated trait information to improve the accuracy of early predictions of breeding values for length of productive life. Proceedings International Workshop on Genetic Improvement of Functional Traits in Cattle, Gembloux, Belgium, January 1996. Department of Animal Breeding and Genetics, SLU. Uppsala, Sweden. Interbull Bull. 1996; 12: 125-135.
Weigel K.A., Lawlor T.J. Adjustment for heterogeneous variance in genetic evaluations for conformation of United States Holsteins. J. Dairy Sci. 1994; 77: 1691-1701.
Wiggans G.R. NCDHIP participation as of January 1, 1997. National Cooperative Dairy Herd Improvement Program Handbook, Fact Sheet K-1. Extension Service, U.S. Department of Agriculture. Washington, DC. 1997a. [http://aipl.arsusda.gov/publish/dhi/dhi97/97k1.html].
Wiggans G.R. USDA summary of 1996 herd averages. National Cooperative Dairy Herd Improvement Program Handbook, Fact Sheet K-3. Extension Service, U.S. Department of Agriculture. Washington, DC. 1997b. [http://aipl.arsusda.gov/publish/dhi/dhi97/97k3.html].
Wiggans G.R., Misztal I., Van Vleck L.D. Implementation of an animal model for genetic evaluation of dairy cattle in the United States. J. Dairy Sci. 1988; 71(Suppl. 2): 54-69.
Wiggans G.R., VanRaden P.M. Effect of including parity-age classes on estimated genetic trend for milk and component yields. J. Dairy Sci. 1994; 77(Suppl. 1): 267 (Abstract).
Wiggans G.R., VanRaden P.M. Flow of information for genetic evaluation of yield traits. Proceedings of the Symposium on Continuous Evaluation in Dairy Cattle, College Park, Maryland, June 13, 1993. Misztal, I. (ed.). Department of Animal Sciences, University of Illinois. Urbana, Illinois. Pages 19-29. 1993.
Wiggans G.R., VanRaden P.M. Including information from records in later herds in animal model evaluations. J. Dairy Sci. 1990; 73: 3336-3339.
Wiggans G.R., VanRaden P.M. Method and effect of adjustment for heterogeneous variance. J. Dairy Sci. 1991; 74: 4350-4357.
Wiggans G.R., VanRaden P.M. USDA-DHIA animal model genetic evaluations. National Cooperative Dairy Herd Improvement Program Handbook, Fact Sheet H-2. Extension Service, U.S. Department of Agriculture. Washington, DC. 1989. [http://www.inform.umd.edu/EdRes/Topic/AgrEnv/ndd/dairy/ANIMAL_MODEL_GENETIC_EVALUATIONS.html].
Wiggans G.R., VanRaden P.M., Powell R.L. A method for combining United States and Canadian bull evaluations. J. Dairy Sci. 1992; 75: 2834-2839.
Wiggans G.R., VanRaden P.M., Zuurbier J. Calculation and use of inbreeding coefficients for genetic evaluation of United States dairy cattle. J. Dairy Sci. 1995; 78: 1584-1590.